Mathematical Biology

My research interests involve using the techniques of mathematics to explore how the laws of physics inform biological function. As a general rule, this means that I design mathematical models to describe biological systems of interest, then develop the mathematical tools necessary to solve, analyze, or simulate said models. Finally, these mathematical results are reinterpreted in a biological context in order to gain insight into how cells, tissues, and organisms operate.
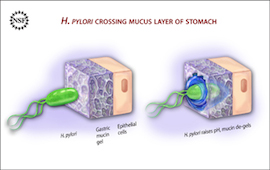
I am particularly interested in the mechanics of fluids and complex fluids, and transport processes therein, all of which are important in biological and physiological systems. My mathematical training focuses on techniques for solving and analyzing partial differential equations (PDEs), as well as numerical methods for complex materials. I have worked on developing an Immersed Boundary (IB) method for poro-elastic materials in the context of cellular modelling, as well as integration of electro-diffusion equations into models of viscous two-phase media (in the context of gastric mucus). I have also developed models of strain relaxation in filamentous actin networks in emmulsion droplets.
For a list of my published work and a complete C.V., please see my publications page.